```{r}
#| label: setup
#| message: false
library(tidyverse)
library(janitor)
```3 Billboard Cleaning

“If you’re doing data analysis every day, the time it takes to learn a programming language pays off pretty quickly because you can automate more and more of what you do.” – Hadley Wickham, chief scientist at Posit
3.1 Learning goals for this chapter
- Practice organized project setup.
- Learn a little about data types available to R.
- Learn how to download and import CSV files using the readr package.
- Introduce the tibble/data frame.
- String functions together with the pipe:
|>(or%>%). - Learn how to modify data types (date) and
select()columns.
Every project in this book is built around a data source that can birth acts of journalism. For this first one we’ll be exploring the Billboard Hot 100 music charts. You’ll use R skills to find the answer to a bunch of questions in the data and then write about it.
3.2 Basic steps of this lesson
Before we get into our storytelling, we have to set up our project, get our data and make sure it is in good shape for analysis. This is pretty standard for any new project. Here are the major steps we’ll cover in detail for this lesson (and many more to come):
- Create your project structure
- Find the data and (usually) get it onto on your computer
- Import the data into your project
- Clean up column names and data types
- Export cleaned data for later analysis
So this chapter is about collecting and cleaning data. We’ll handle the analysis separately in another chapter. I usually break up projects this way (cleaning vs analyzing) to stay organized and to avoid repeating the same cleaning steps.
3.3 Create a new project
You learned out to create a new Quarto website in the last chapter so you got this! Here are the basic steps for you to follow:
- Launch RStudio if you haven’t already.
- Make sure you don’t have an existing project open. Use File > Close project if you do.
- Use the
+Rbutton to create a project in a New Directory. For project type, choose Quarto Website. - Name the project
yourfirstname-billboardand put it in yourrwd/folder.
3.3.1 Update index with project description
For our projects, we want to use the index.qmd file (and our resulting website home page) as a description of the project. This is so you, your editor and anyone you share this with can see what this is about.
To save time and such, I’ll supply you with a short project description.
---
title: "Billboard Hot 100"
---
This project is an assignment in the Reporting with Data class at the University of Texas at Austin.
It is a study of [Billboard Hot 100](https://www.billboard.com/charts/hot-100/) chart data as collected and updated by Christian McDonald in a [Github repo](https://github.com/utdata/rwd-billboard-data). While the archive was compiled through a number of methods, the original source is Billboard Magazine, owned by the Penske Media Corporation. The data dates back to the Hot 100's beginning in August 1958 and is updated weekly, but the end date here will depend on when the user runs the code.- Copy the text above and replace the contents of
index.qmd. - Use the Render button or Cmd-Shift-K to render the page.
You want each of your projects to have a description like this, along with details describing the source data, the goals of the project, the findings and perhaps links to stories and graphics published based on the data analysis.
In the next chapter you’ll return to this page to update it with your findings. For now we’re done with this file so you can close it.
3.3.2 Update our _quarto.yml
The default _quarto.yml configuration file is fine, but there are some adjustments we can make to make our lives easier and our rendered project more beautiful.
We will use copy/paste to replace this whole file with the code I have below, but let me walk through what we are changing and why:
- We’ll add a “render” section that helps specify the order that files are processed. You’ll soon see we will name our files with numbers like
01-to specify this order. We sometimes perform functions in certain notebooks, liked downloading data, that needs to be processed before other files. - For the
title:change the value to"Billboard Hot 100". This is the main website name that displays in the navigation bar. - We’ll change the format of our website navigation to a “sidebar” instead of a “navbar” that runs across the top of the page.
- Sidebars define the content using “content” instead of “left”. We’ll set ours to list every
.qmdpage we create instead of listing them individually. - We are adding a setting to make our rendered data frames prettier with
df-print: paged. - We add one more setting
code-overflow: wrapthat improves the display of our code on rendered pages.
Before you replace the contents, look at what you have in your _quarto.yml file compared to this below so you can recognized the differences.
- Replace the contents of your
_quarto.ymlfile with the contents shown below. - Render your site again so you can see the changes.
_quarto.yml
project:
type: website
render:
- "01*.qmd"
- "*.qmd"
website:
title: "Billboard Hot 100"
sidebar:
contents:
- href: index.qmd
text: Home
- auto: "*.qmd"
format:
html:
theme: cosmo
css: styles.css
toc: true
df-print: paged
code-overflow: wrapThere are many other changes you can make using _quarto.yml configurations, but we won’t get into them here. See the Quarto Guide for more information.
3.3.3 Create directories for your data
Next we will create some data folders in our project. We’ll use standard names for consistency and we’ll do this for every project we build.
The first folder is called data-original. We are creating this folder because we want to keep a pristine version of our original data that we never change or overwrite. This is a core data journalism commandment: Thou shalt not change original data.
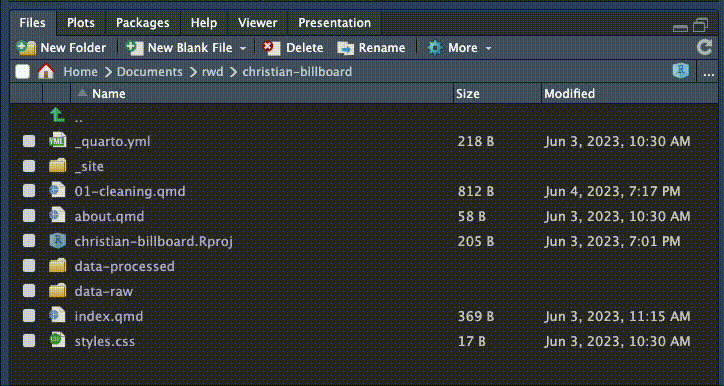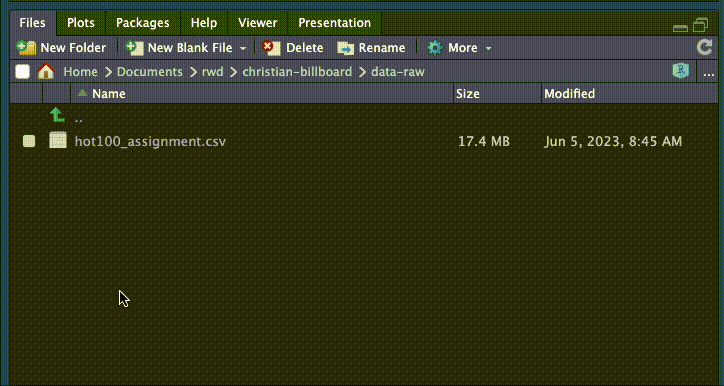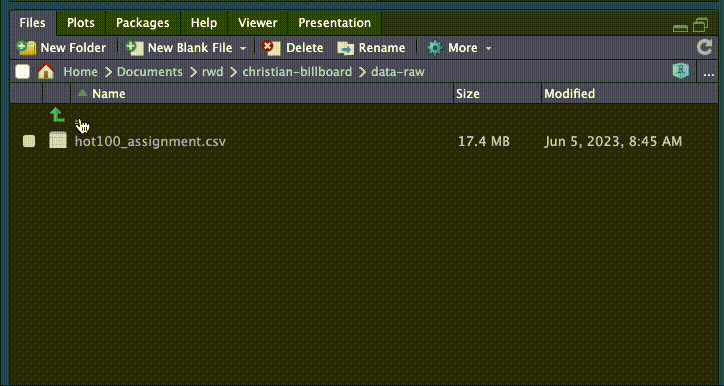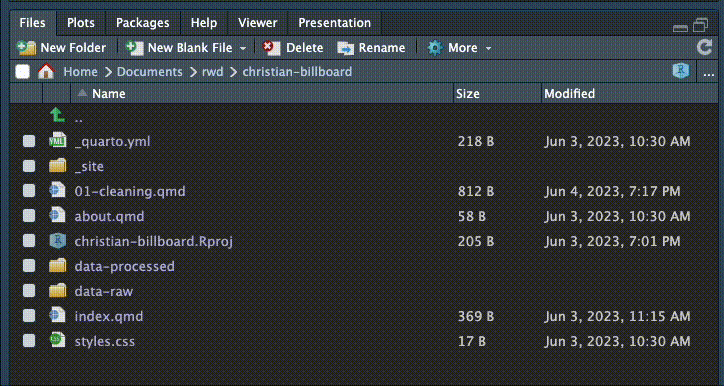
In your Files pane at the bottom-right of RStudio, there is a New Folder icon.
- Click on the New Folder icon.
- Name your new folder
data-original. This is where we’ll put original data from our source that we don’t want to ever overwrite. - Create an additional folder called
data-processed. This is were we will write any data we create.
Once you’ve done that, they should show up in the file explorer in the Files pane. Click the refresh button if you don’t see them. (The circlish thing at top right of the screenshot below. You might have to widen the pane to see it.)
Again, we’ll do this with every new project we create:
- Create a Quarto website
- Update the index with information about the project
- Create
data-originalanddata-processedfolders.
Since you’ll be creating projects multiple times, I’ve saved the basic steps – especially the _quarto.yml contents – in an appendix chapter: Project setup. You can use it as a reference going forward.
3.4 Create our cleaning notebook
We’ll typically use at least three Quarto files in our projects:
- The
index.qmdthat describes the project. - A “Cleaning” notebook where we prepare our data.
- An “Analysis” notebook where we pose and answer our data questions.
So let’s create our “Cleaning” notebook.
- In the bottom right Files pane, click on the New File button (a white box with a green
+sign) and choose the Quarto Document to start a new notebook. - Set the filename to
01-cleaning.qmd. - Set the title to “Cleaning”.
- Save the new file (Do Cmd-S or look under the File menu or use the floppy disc icon in the tool bar.)
We named this notebook starting with 01- because we will eventually end up with multiple notebooks that depend on each other and we will need to know the order to run them in the future.
3.4.1 Describe the goals of the notebook
At the top of the cleaning file after the YAML metadata we’ll want to explain the goals of the notebook and what we are doing.
Add this text to your notebook AFTER the metadata:
## Goals of this notebook The steps we'll take to prepare our data: - Download the data - Import it into our notebook - Clean up data types and columns - Export the data for next notebook
We want to start each notebook with a list like this so our future selves and others know what we are trying to accomplish. It’s not unusual to update this list as we work through the notebook.
We will also write Markdown like this to explain each new “section” or goal as we tackle them.
3.5 The setup chunk
In our previous chapter we installed several R packages onto our computer that we’ll use in almost every lesson. There are others we’ll install and use later.
But to use these packages in our notebook (and the functions they provide us) we have to load them using the library() function. We always have to declare these and convention dictates we put it near the top of a notebook so everyone understands what is needed to run our code.
3.5.1 Load the libraries
- In your notebook after the goals listing, create a new “section” by adding a headline called “Setup” using Markdown, like this:
## Setup. - After the. headline use Cmd+option+i to insert an R code chunk.
- Inside that chunk, I want you to type the code shown below.
I want you to type the code so you can see how RStudio helps you complete the code. It’s something you have to see or do yourself to understand how RStudio helps you type commands, but as you type RStudio will give you valid options you can scroll through and hit tab key to choose.
Here is a gif of me typing in the commands. I’m using keyboard commands like the up and down arrow to make selections, and the tab key to select them. This concept is called code completion.
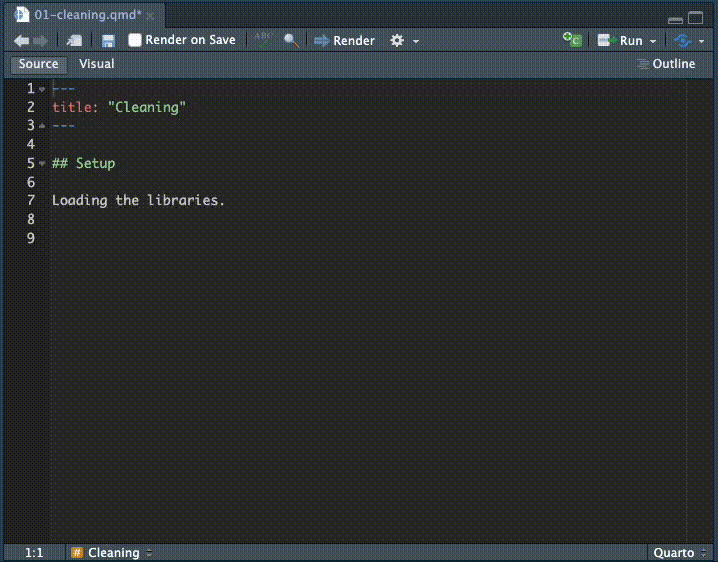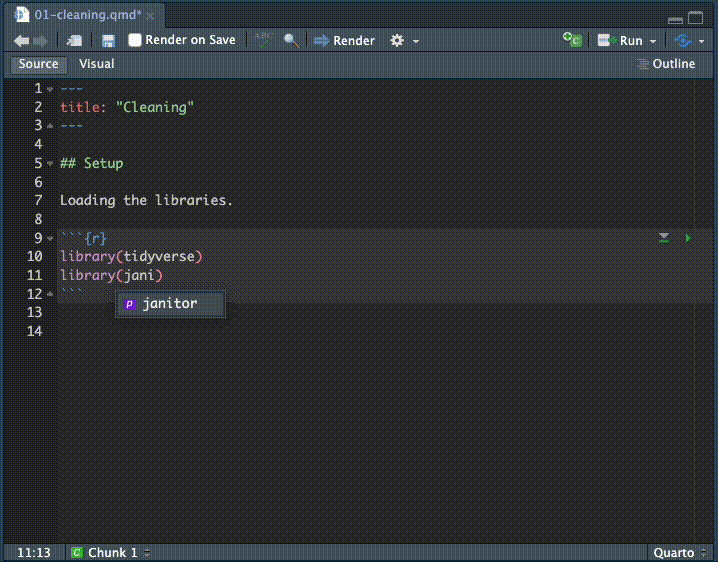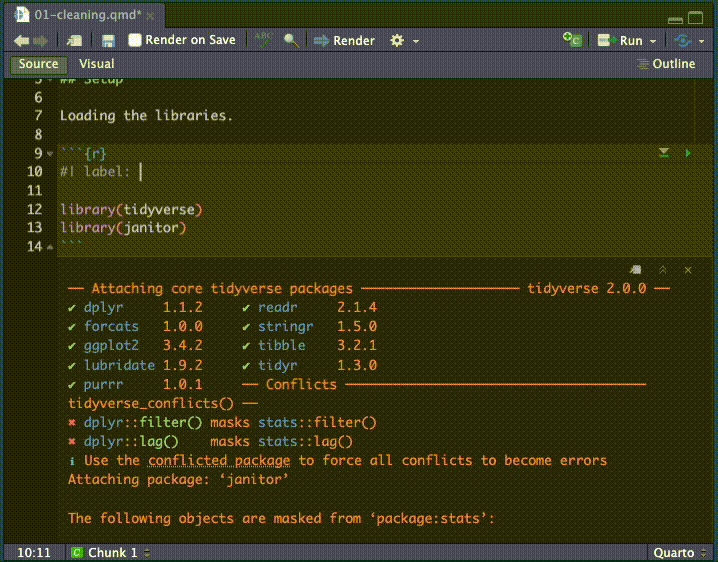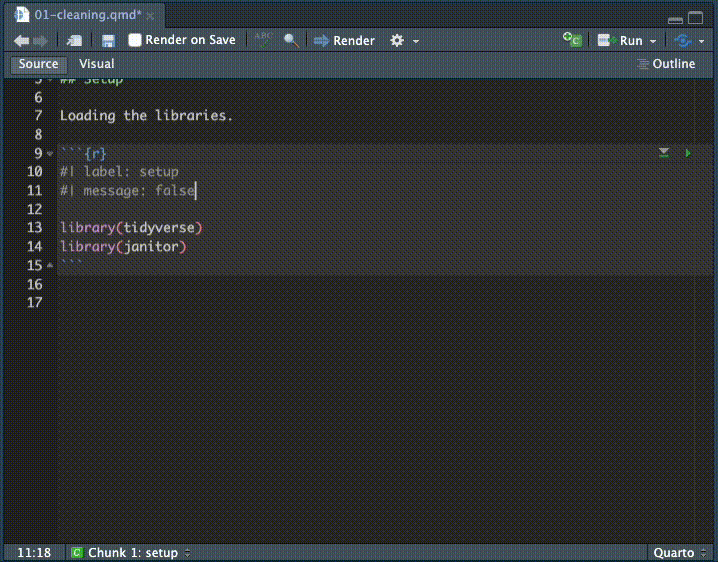
Here is the code with the chunk bits, etc:
3.5.2 About the libraries
A little more about the two packages we are loading here. We use them a lot.
- The
tidyversepackage is actually a collection of packages for data science that are designed to work together. You can see the first time I run the chunk in the gif above that a bunch of libraries were loaded. By loading the whole tidyverse library we getreadrfunctions for importing data,dplyrto manipulate data,lubridateto help work with dates, andggplotto visualize data. There are more. - The
janitorpackage is not maintained by Posit but it has many useful functions to clean and view data. I always include it. We’ll use janitor’sclean_names()later.
3.5.3 About the options
After adding the libraries I go back and add some lines at the top of the chunk called execution options. Let’s talk about each line:
- We start with
#| label: setupwhich is an execution option to “name” the code chunk. Labeling chunks is optional but useful.- The chunk label
setupis special as it will be run first if it hasn’t already been run. - When we name a code chunk it creates a bookmark of sorts, which I’ll show you later.
- The chunk label
- The
#| message: falseoption suppress the message about all the packages inside tidyverse. We don’t need to display that in our notebook so I usually only add that for my setup chunk to suppress them.
We’ve done a lot of work setting up our project, and we’re about to get into some important info about our the data we’ll be working with. This would be a good place to take a break if you need to. Seriously, go eat a cookie 🍪.
3.6 About the Billboard Hot 100
The Billboard Hot 100 singles charts has been the music industry’s standard record chart since its inception on Aug. 4th, 1958. The rankings, published by Billboard Magazine, are currently based on sales (physical and digital), radio play, and online streaming. The methods and policies of the chart have changed over time.
The data we will use was compiled by Prof. McDonald from a number of sources. When you write about this data (and you will), you should source it as the Billboard Hot 100 from Billboard Magazine, since that is where it originally came from and they are the “owner” of the data.
3.6.1 Data dictionary
Take a look at the current chart. Our data contains many (but not quite all) of the elements you see there. Each row of data (or observation as they are known in R) represents a song and the corresponding position on that week’s chart. Included in each row are the following columns (a.k.a. variables):
- CHART DATE: The release date of the chart
- THIS WEEK: The current ranking as of the chart date
- TITLE: The song title
- PERFORMER: The performer of the song
- LAST WEEK: The ranking on the previous week’s chart
- PEAK POS.: The peak rank the song has reached as of the chart date
- WKS ON CHART: The number of weeks the song has appeared as of the chart date
3.6.2 Let’s download our data
Our data is stored on a code sharing website called Github, and it is formatted as a “comma-separated value” file, or .csv. That means it basically a text file where every line is a new row of data, and each field is separated by a comma.
Because it is on the internet and has a URL, we could import it directly into our project from there, but there are benefits to getting the file onto your computer first. The hosted file could change later in ways you can’t control, and you would need Internet access to the file the next time you ran your notebook. If you download the file to keep your own copy, you have more control of your future.
Since this is a new “section” of our cleaning notebook, we’ll note what we are doing and why in Markdown.
- Add a Markdown headline
## Downloading dataand some text explaining you are downloading data. Include a note about where the data comes from and include a link to the original source. (This way others and future self will know where the data came from.) - Create an R chunk and copy/paste the following inside it. (Given the long URL, go ahead and use the copy icon at the top right of the chunk):
download.file(
"https://github.com/utdata/rwd-billboard-data/blob/main/data-out/hot100_assignment.csv?raw=true",
"data-original/hot100_assignment.csv",
mode = "wb"
)Let’s explain about this:
This download.file() code is a function, or a bit of code that has been written to do a specific task. This one (surprise!) downloads files from the internet. As a function, it has some “arguments”, which are expected pieces of information that the function needs to do its job. Sometimes we just enter the arguments in the order that the function expects them, like we do here. Other times we will give it the kind of argument like mode = "w". You can search for documentation about functions in the Help tab in the bottom-right pane of RStudio.
In this case, we are supplying three arguments:
- The first is the URL of the file you are downloading. Note that this is in quotes.
- The second is the location where we want to save the file, and what want to name it. We call this a “path” and you can see it looks a lot like a URL. This path is in relation to the document we are in, so we are saying we want to put this file in the
data-original/folder with a name ofhot100_assignment.csv, but it is really one path:data-original/hot100_assignment.csv. - The third argument
mode = "w"gets into the weeds a little, but it helps Windows computers understand the file.
When you run the function you get an output similar to this:
trying URL 'https://github.com/utdata/rwd-billboard-data/blob/main/data-out/hot100_assignment.csv?raw=true'
Content type 'text/plain; charset=utf-8' length 18924862 bytes (18.0 MB)
==================================================
downloaded 18.0 MBThat’s not a small file at about 18 MB and 300,000+ rows, but it’s not a huge one, either.
You can check that this worked by going to your Files tab and clicking on the data-original folder to go inside it and see if the file is there. To return out of the folder, click on the two dots to the right of the green up arrow.
3.7 Import the data
Screenshots, gifs and code results in this book may differ slightly from your project since you are using newer data!
Now that we have the data on our computer, let’s import it into our notebook so we can see it.
Since we are doing a new thing, we should again note what we are doing.
- Add a Markdown headline:
## Import data - Add some text to explain that we are importing the Billboard Hot 100 data.
- After your description, add a new code chunk (Cmd+Option+i).
We’ll be using the read_csv() function from the tidyverse readr package, which is different from read.csv that comes with base R. read_csv() is mo betta.
Inside the function we put in the path to our data, inside quotes. If you start typing in that path and hit tab, it will complete the path. (Easier to show than explain).
- Add the following code into your chunk and run it.
read_csv("data-original/hot100_assignment.csv")Note the path to the file is in quotes!
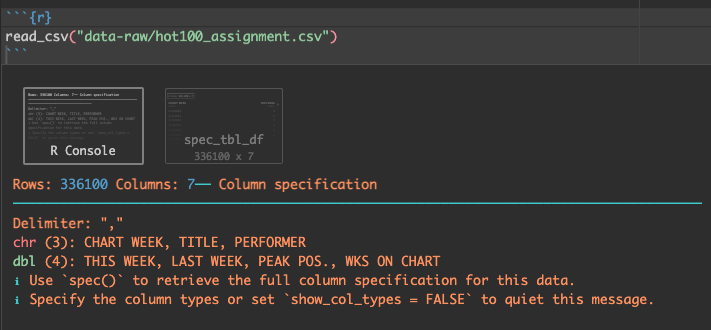
You get two results printed to your notebook.
The first result called “R Console” shows what columns were imported and the data types. It’s important to review these to make sure things happened the way that expected. In this case it noted which columns came in as text (chr), or numbers (dbl). The red colored text in this output is NOT an indication of a problem.
The second result spec_tbl_df prints out the data like a table. The data object is a tibble, which is a fancy tidyverse version of a “data frame”.
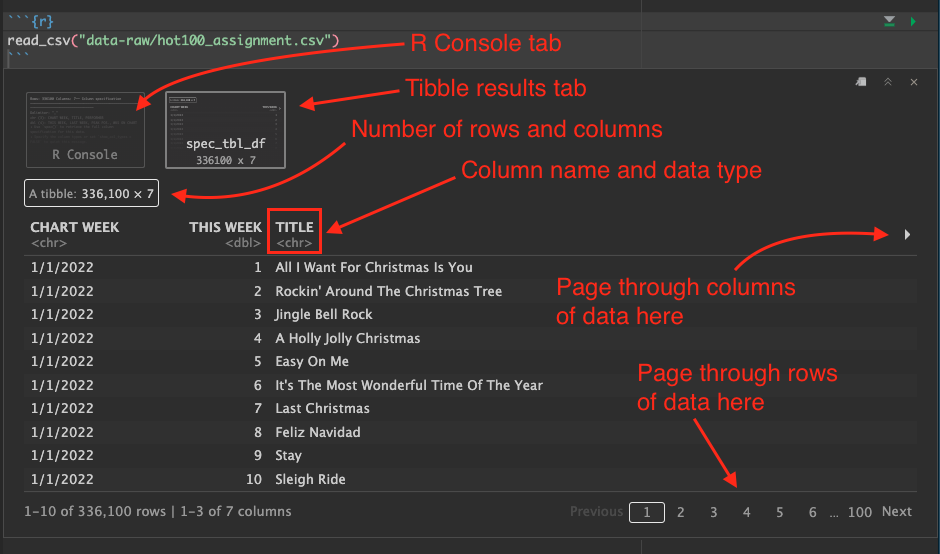
When we look at the data output in RStudio, there are several things to note:
- Below each column name is an indication of the data type. This is important.
- You can use the arrow icon on the right to page through the additional columns.
- You can use the paging numbers and controls at the bottom to page through the rows of data.
- The number of rows and columns is displayed.
At this point we have only printed this data to the screen. We have not saved it in any way, but that is next.
I will use the term tibble and data frame interchangeably. Think of tibbles and data frames like a well-structured spreadsheet. They are organized rows of data (called observations) with columns (called variables) where every column is a specific data type.
3.7.1 Assign our import to an R object
Now that we know how to find our data, we next need to assign it to an R object so it can be a named thing we can reuse. We don’t want to re-import the data over and over each time we need it.
The syntax to create an object in R can seem weird at first, but the convention is to name the object first, then insert stuff into it. So, to create an object, the structure is this:
# this is pseudo code. Don't put it in your notebook.
new_object <- stuff_going_into_objectThink of it like this: You must have a bucket before you can fill it with water. We “name” the bucket, then fill it with data. That bucket is then saved into our “environment”, meaning it is in memory where we can access it easily by calling its name.
Let’s make a object called hot100_raw and fill it with our imported tibble.
- Edit your existing code chunk to look like below. You can add the
<-by using Option+- as in holding down the Option key and then pressing the hyphen:
hot100_raw <- read_csv("data-original/hot100_assignment.csv")Run that chunk and several things happen:
- We no longer see the result of the data in the notebook because we created an object instead of printing it.
- In the Environment tab at the top-right of RStudio, you’ll see the
hot100_rawobject listed.- Click on the blue play button next to
hot100_rawobject and it will expand to show you a summary of the columns. - Click on the name
hot100_rawand it will open a “View” of the data in another window, so you can look at it in spreadsheet form. You can even sort and filter it.
- Click on the blue play button next to
- Once you’ve looked at the data, close the data view by clicking the little
xnext to the tab name.
3.7.2 Print a peek to your R Notebook
Since we can’t see the data after we assign it, let’s print the object to our notebook so we can refer to it.
- Edit your import chunk to add the last two lines of this, including
#:
# create the object, then fill it with data from the csv
hot100_raw <- read_csv("data-original/hot100_assignment.csv")
# peek at the data
hot100_rawYou can use the green play button at the right of the chunk, or preferrably have your cursor inside the chunk and do Cmd+Shift+Return to run all lines. (Cmd+Return runs only the current line.)
3.7.3 Glimpse the data
There is another way to peek at the data that I use a lot because it is more compact and shows you all the columns and data examples without scrolling: glimpse().
- After your last code chunk, add a sentence that says you will print a “glimpse” of your data.
- Create a new code chunk and add the code noted below.
I’m showing the return here as well. Afterward I’ll explain the pipe: |>.
# glimpse the data
hot100_raw |> glimpse()Rows: 352,300
Columns: 7
$ `CHART WEEK` <chr> "1/1/2022", "1/1/2022", "1/1/2022", "1/1/2022", "1/1/20…
$ `THIS WEEK` <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, …
$ TITLE <chr> "All I Want For Christmas Is You", "Rockin' Around The …
$ PERFORMER <chr> "Mariah Carey", "Brenda Lee", "Bobby Helms", "Burl Ives…
$ `LAST WEEK` <dbl> 1, 2, 4, 5, 3, 7, 9, 11, 6, 13, 15, 17, 18, 0, 8, 25, 1…
$ `PEAK POS.` <dbl> 1, 2, 3, 4, 1, 5, 7, 6, 1, 10, 11, 8, 12, 14, 7, 16, 12…
$ `WKS ON CHART` <dbl> 50, 44, 41, 25, 11, 26, 24, 19, 24, 15, 31, 18, 14, 1, …The glimpse function shows there are 300,000+ rows and 7 columns in our data. Each column is then listed out with its data type and the first several values in that column.
3.7.4 About the pipe |>
We need to break down this code a little: hot100_raw |> glimpse().
We are starting with the object hot100_raw, but then we follow it with |>, which is called a pipe. The pipe is a construct that takes the result of an object or function and passes it into another function. Think of it like a sentence that says “AND THEN” the next thing.
Like this:
I woke up |>
got out of bed |>
dragged a comb across my headIf you are breaking your code into multiple lines using pipes, then the |> needs to be at the end of a line and the next line should be indented so there is a visual clue it is related to line above it, like this:
hot100_raw |>
glimpse()It might look like there are no arguments inside glimpse(), but what we are actually doing is passing the hot100_raw tibble into it like this: glimpse(hot100_raw). For almost every function in R the first argument is “what data are you taking about?” The pipe allows us to say “hey, take the data we just mucked with (i.e., the code before the pipe) and use that in this function.”
There is a keyboard command for the pipe |>: Cmd+Shift+m. Learn that one!
A rabbit dives into a pipe
The concept of the pipe was first introduced by tidyverse developers in 2014 in a package called magrittr. They used the symbol %>% as the pipe. It was so well received the concept was written directly into base R in 2021, but using the symbol |>. When Hadley Wickham’s updated R for Data Science in 2021 he started using the base R pipe |> so I am, too. You can configure which to use in RStudio.
This switch to |> is quite recent so you might see %>% used in this book. Assume |> and %>% are interchangeable. There is A LOT of code in the wild using the magrittr pipe %>%, so you’ll find many references on Stack Overflow and elsewhere.
3.8 Cleaning data
Data is dirty. Usually because a human was involved at some point, and we humans are fallible.
Data problems are often revealed when importing so it is good practice to check for problems and fix them right away. We’ll face some of those challenges in this project, but we should talk about what is good vs dirty data.
Good data should:
- Have a single header row with well-formed column names.
- Descriptive names are better than not descriptive.
- Short names are better than long ones.
- Spaces in names make them harder to work with. Use
_or.between words. I prefer_and all lowercase text.
- Each column should have the same kind of data: numbers vs words, etc.
- Remove notes or comments from data files.
3.8.1 Cleaning column names
So, given those notes above, we should clean up our column names. This is why we have included the janitor package, which includes a neat function called clean_names()
- Edit your import chunk to add a pipe and the clean_names function:
|> clean_names() - Rerun both the import chunk and the glimpse chunk!
hot100_raw <- read_csv("data-original/hot100_assignment.csv") |> clean_names()
# peek at the data
hot100_rawhot100_raw |> glimpse()Rows: 352,300
Columns: 7
$ chart_week <chr> "1/1/2022", "1/1/2022", "1/1/2022", "1/1/2022", "1/1/2022…
$ this_week <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17…
$ title <chr> "All I Want For Christmas Is You", "Rockin' Around The Ch…
$ performer <chr> "Mariah Carey", "Brenda Lee", "Bobby Helms", "Burl Ives",…
$ last_week <dbl> 1, 2, 4, 5, 3, 7, 9, 11, 6, 13, 15, 17, 18, 0, 8, 25, 19,…
$ peak_pos <dbl> 1, 2, 3, 4, 1, 5, 7, 6, 1, 10, 11, 8, 12, 14, 7, 16, 12, …
$ wks_on_chart <dbl> 50, 44, 41, 25, 11, 26, 24, 19, 24, 15, 31, 18, 14, 1, 49…The clean_names() function has adjusted all the variable names, making them all lowercase and using _ instead of periods between words. Believe me when I say this is helpful when you are writing code. It makes type-assist work better and you can now double-click on a column name to select all of it and copy and paste somewhere else. When you have spaces, dots or dashes in an object you can’t double-click on it to select all of it.
3.8.2 Fixing the date
Dates in programming are a tricky because they are evaluated under the hood as “the number of seconds before/after January 1, 1970”. Yes, that’s crazy, but it is also cool because that allows us to do math on them. So, to use our chart_date properly in R we want to convert it from text datatype into a real date datatype. (If you wish, you can read more about why dates are tough in programming | PDF version.)
Converting text into dates can be challenging, but the tidyverse universe has a package called lubridate to ease the friction. (Get it?).
Since we are doing something new, we want to start a new section in our notebook and explain what we are doing.
- In Markdown add a headline:
## Fix our dates. - Add some text that you are using lubridate to create a new column with a real date.
- Add a new code chunk. Remember Cmd+Option+i will do that.
We will be changing or creating our data, so we will create a new object to store it in. We do this so we can go back and reference the unchanged data if we need to. Because of this, we’ll set up a chunk of code that allows us to peek at what is happening while we write our code. We’ll do this kind of setup often when we are working out how to do something in code.
- Add the following inside your code chunk.
- Run the code, and then read through the annotations below.
Below is our first use of annotated code with the circled numbers shown on the right edge of the code block. Those numbers correspond with the explanation under the code chunk. The best way to use code in chunks like these is to a) type it yourself (preferred!) or b) to hover you cursor over the code chunk to expose the “Copy to clipboard” icon that you can use to copy the code and then paste it into your notebook.
What you can’t do is highlight all the text in the code chunk and then paste it. That will include the numbers and the code will break.
1# part we will build upon
2hot100_date <- hot100_raw
3
4# peek at the result
5hot100_date |> glimpse()- 1
-
I have a comment starting with
#to explain the first part of the code. - 2
-
We created a new object (or “bucket”) called
hot100_dateand we fill it with ourhot100_rawdata. Yes, as of now they are exactly the same. - 3
- I leave a blank line between the two lines of code for clarity …
- 4
- Then another optional comment explaining the code …
- 5
-
Then we take the new
hot100_dateobject and pipe it intoglimpse()so we can see changes as we work.
Rows: 352,300
Columns: 7
$ chart_week <chr> "1/1/2022", "1/1/2022", "1/1/2022", "1/1/2022", "1/1/2022…
$ this_week <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17…
$ title <chr> "All I Want For Christmas Is You", "Rockin' Around The Ch…
$ performer <chr> "Mariah Carey", "Brenda Lee", "Bobby Helms", "Burl Ives",…
$ last_week <dbl> 1, 2, 4, 5, 3, 7, 9, 11, 6, 13, 15, 17, 18, 0, 8, 25, 19,…
$ peak_pos <dbl> 1, 2, 3, 4, 1, 5, 7, 6, 1, 10, 11, 8, 12, 14, 7, 16, 12, …
$ wks_on_chart <dbl> 50, 44, 41, 25, 11, 26, 24, 19, 24, 15, 31, 18, 14, 1, 49…To be clear, we haven’t changed any data yet. We just created a new object exactly like the old object.
3.8.2.1 Working with mutate()
We are going to use the text inside chart_date field to create a new “real” date. We will use the dplyr function mutate() to do this, with some help from lubridate.
dplyr is the tidyverse package of functions to manipulate data. We’ll use functions from it a lot. Dplyr is loaded with the library(tidyverse) so you don’t have to load it separately.
Let’s explain how mutate() works first: The function mutate() changes data within a column of data based on the rules you feed it. You can either create a new column or overwrite an existing one.
Within the mutate() function, we name the changed thing first (the bucket!) and then fill it with the new mutated value.
# This is just explanatory psuedo code
# You don't need this in your notebook
data |>
mutate(
newcol = new_stuff_from_math_or_whatever
)That new value could be arrived at through math or any combination of other functions. In our case, we want to convert our old text-based date to a real date, and then assign it back to the “new” column.
- Edit your chunk to add the changes below and run it. I implore you to type the changes so you see how RStudio helps you write it. Use tab completion, etc.
- 1
-
At the end of the first line, we added the pipe
|>because we are taking ourhot100data AND THEN we will mutate it. (Remember Cmd-shift-m for the pipe!) - 2
-
Next, we start the
mutate()function. If you use type assist and tab completion to type this in, your cursor will end up in the middle of the parenthesis. This allows you to then hit your Return key to split it into multiple lines with proper indenting. We do this so we can more clearly see what inside the mutate … where the real action is going on here. It’s also possible to make multiple changes within the same mutate function, and putting each one on their own line makes that more clear. - 3
-
Inside the mutate, we first name our new column
chart_dateand then we set that equal tomdy(chart_week), which is explained next.
The mdy() function is part of the lubridate package. Lubridate allows us to parse text and then turn it into a real date if we tell it the order of the date values in the original data.
- Our original date was something like “7/17/1965”. That is month, followed by day, followed by year.
- We use the lubridate function
mdy()to say “that’s the order this text is in, now please convert this into a real date”, which shows as YYYY-MM-DD. Lubridate is smart enough to figure out if you have/or-between your values in the original text.
If your original text is in a different date order, then you look up what lubridate function you need to convert it. I typically use the cheatsheet in the lubridate documentation. You’ll find them in the PARSE DATE-TIMES section.
Good coding style is like correct punctuation: you can manage without it, butitsuremakesthingseasiertoread.
Programming languages often have a “syntax” of conventions that people follow about how you indent code, where you put spaces and how you name things. We follow the Tidyverse Style Guide for R, and you should to. RStudio will help you indent properly as you type. (Easier to show than explain.)
I also often use strategically placed returns to make the code more readable. The code above would work the same if it were all on the same line, but writing it this way helps me understand it. When I do this, I’m usually putting each argument of a function on its own line, as noted in the Pipes chapter of the style guide.
3.8.2.2 Check the result!
This new chart_date column is added as the LAST column of our data. After doing any kind of mutate you want to check the result to make sure you got the results you expected, which is why we didn’t just overwrite the original chart_week column. That’s also why we built our code this way with glimpse() so we can see example of our data from both the first and the last column. (We’ll rearrange all the columns in a bit once we are done cleaning everything.)
Check your glimpse returns … did your dates convert correctly?
3.8.3 Arrange the data
Just to be tidy, we want ensure our data is arranged to start with the oldest week and “top” of the chart and then work forward through time and rank.
i.e., let’s arrange this data so that the oldest data is at the top.
Sorting data is not a particularly difficult concept to grasp, but it is one of the Basic Data Journalism Functions, so watch this video:
In R, the function we use to sort our data is arrange().
We’ll build upon our existing code and use the pipe |> to push it into an arrange() function. Inside arrange we’ll feed it the columns we wish to sort by.
- Edit your chunk to the following to add the
arrange()function:
# part we will build upon
hot100_date <- hot100_raw |>
mutate(
chart_date = mdy(chart_week)
1 ) |>
2 arrange(chart_date, this_week)
# peek at the result
hot100_date |> glimpse()- 1
- Add the pipe to the end of this line …
- 2
-
… and the arrange line. We list the
chart_datecolumn first so we get the earliest charts the top, then thethis_weekcolumn so they will be listed 1 through 100.
Rows: 352,300
Columns: 8
$ chart_week <chr> "8/4/1958", "8/4/1958", "8/4/1958", "8/4/1958", "8/4/1958…
$ this_week <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17…
$ title <chr> "Poor Little Fool", "Patricia", "Splish Splash", "Hard He…
$ performer <chr> "Ricky Nelson", "Perez Prado And His Orchestra", "Bobby D…
$ last_week <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ peak_pos <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17…
$ wks_on_chart <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ chart_date <date> 1958-08-04, 1958-08-04, 1958-08-04, 1958-08-04, 1958-08-…Now when you look at the glimpse, the first record in the chart_date column is from “1958-08-04” and the first in the this_week is “1”, which is the top of the chart.
Just to see this all clearly in table form, we’ll print the top of the table to our screen so we can see it.
- Add a line of Markdown text in your notebook explaining your are looking at the table.
- Add a new code chunk and add the following.
hot100_date |> head(10)- Use the arrows to look at the other columns of the data (which you can’t see in the book).
The head() function prints the first few lines of data. You can choose a specific number of lines by adding a number as the first argument, as we do with “10” here.
It’s OK that the last_week columns has “NA” for those first rows because this is the first week ever for the chart. There was no last_week.
3.8.3.1 Getting summary stats
Printing your data to the notebook can only tell you so much. Yes, you can arrange by different columns to see the maximum and minimum values, but it’s hard to get an overall sense of your data that way when there is 300,000 rows like we have here. Luckily there is a nice function called summary() that gives you some summary statistics for each column.
- Add some Markdown text that you’ll print summary stats of your data.
- Add a new R chunk and put the following in and run it.
hot100_date |> summary() chart_week this_week title performer
Length:352300 Min. : 1.0 Length:352300 Length:352300
Class :character 1st Qu.: 26.0 Class :character Class :character
Mode :character Median : 51.0 Mode :character Mode :character
Mean : 50.5
3rd Qu.: 75.0
Max. :100.0
last_week peak_pos wks_on_chart chart_date
Min. : 0 Min. : 1.00 Min. : 1.000 Min. :1958-08-04
1st Qu.: 22 1st Qu.: 13.00 1st Qu.: 4.000 1st Qu.:1975-06-21
Median : 46 Median : 37.00 Median : 7.000 Median :1992-05-09
Mean : 47 Mean : 40.44 Mean : 9.443 Mean :1992-05-08
3rd Qu.: 71 3rd Qu.: 65.00 3rd Qu.: 13.000 3rd Qu.:2009-03-28
Max. :100 Max. :100.00 Max. :112.000 Max. :2026-02-07
NA's :32460 These summary statistics can be informative for us. It is probably the easiest way to check what the newest and oldest dates are in your data (see the Min. and Max. returns for chart_date). You get an average (mean) and median for each number, too. You might notice potential problems in your data, like if we had a this_week number higher than “100” (we don’t).
3.8.3.2 Note the max date
Since this data could update each week, it is a good idea to note in your notebook what is the most recent chart date. You can find that as the Max. value for chart_date in your summary. You’ll need this value when you write your story.
3.8.4 Selecting columns
Now that we have the fixed date column, we don’t need the old chart_week version that is text. We’ll use this opportunity to discuss select(), which is another concept in our Basic Data Journalism Functions series, so watch this:
In R, the workhorse of the select concept is the function called — you guessed it — select(). In short, the function allows us to choose a subset of our columns. We can either list the ones we want to keep or the ones we don’t want.
Like a lot of things in R, select() is both easy and complex. It’s really easy to just list the columns you want to keep. But select() can also be very powerful as you learn more options.
We’ll add one bit of complexity: We’ll also rename some columns as we list them.
- Add a Markdown headline:
## Selecting columns. - Explain in text we are dropping the text date column and renaming others.
- Add the code below and then I’ll explain it. We again are setting this up to create a new object and view the changes.
1hot100_clean <- hot100_date |>
2 select(
3 chart_date,
4 current_rank = this_week,
title,
performer,
previous_rank = last_week,
peak_rank = peak_pos,
wks_on_chart
)
5hot100_clean |> glimpse()- 1
-
Name our new object
hot100_cleanand fill it with the result ofhot100_dateand then include the modifications that follow. - 2
-
We start the
select()function to list the columns to keep - 3
-
We start with
chart_datewhich is our real date. We just won’t ever name the text version so we won’t keep it. - 4
-
When we get to our
this_weekcolumn, we rename it tocurrent_rank. We’re doing this because we’ll rename all the “ranking” columns with something that includes_rankat the end. (While this is good practice, the reasons get into the weeds). - 5
- Lastly, we glimpse the new object to check it.
Rows: 352,300
Columns: 7
$ chart_date <date> 1958-08-04, 1958-08-04, 1958-08-04, 1958-08-04, 1958-08…
$ current_rank <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 1…
$ title <chr> "Poor Little Fool", "Patricia", "Splish Splash", "Hard H…
$ performer <chr> "Ricky Nelson", "Perez Prado And His Orchestra", "Bobby …
$ previous_rank <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ peak_rank <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 1…
$ wks_on_chart <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…There are other ways to accomplish the same thing, but this works for us.
3.9 Exporting data
3.9.1 Using multiple notebooks
It is good practice to separate the cleaning of your data from your analysis. By doing all our cleaning at once and exporting it, we can use that cleaned data over and over in future notebooks.
Because each notebook needs to be self-contained to render, we will export our cleaned data in a native R format called .rds. (Maybe this stands for R data storage?). This format preserves all our data types so we won’t have to reset or clean them.
This is one of the reasons I had you name this notebook 01-cleaning.qmd with a 01 at the beginning, so we know to run this one before we can use the notebook 02-analysis.qmd (next lesson!). I use 01- instead of just 1- in case there are more than nine notebooks. I want them to appear in order in my files directory. I’m anal retentive like that, which is a good trait for a data journalist.
One last thought to belabor the point: Separating your cleaning can save time. I’ve had cleaning notebooks that took 20 minutes to process. Imagine if I had to run that every time I wanted to rebuild my analysis notebook. Instead, the import notebook spits out a clean file that can be imported in a fraction of that time.
This was all a long-winded way of saying we are going to export our data now.
We will use another function from the readr package called write_rds() to create our exported file. We’ll save the data into the data-processed folder we created earlier. We are avoiding our data-original folder because “Thou shalt not change original data” even by accident.
- Create a Markdown headline
## Exportsand write a description that you are exporting files to .rds. - Add a new code chunk and add the following code:
- 1
-
Here we start with the
hot100_cleantibble that we created earlier, and then … - 2
-
We then pipe
|>the result of that into a new functionwrite_rds(). In addition to the data, the function needs the path to where we want to save the file, so in quotes we give it the path to where we want to save the file:"data-processed/01-hot100.rds".
Remember, we are saving into the data-processed folder because we never export into data-original. We are naming the file starting with 01- to indicate to our future selves that this output came from our first notebook. We then name it, and use the .rds extension.
3.9.2 About paths
When I say “path to where we want to save the file” what I’m talking about is the folder structure on your computer. The frame of reference is where your notebook is stored, which is typically in your project folder. When we want to reference other files from within our notebook, we have to provide the path (or folder structure) to where that file is, including its name. We use / in the path to designate we are going inside a folder.
Here is the file structure inside your project folder:
├── 01-cleaning.qmd
├── 02-analysis.qmd
├── _quarto.yml
├── christian-billboard.Rproj
├── data-processed
│ └── 01-hot100.rds
├── data-original
│ └── hot100_assignment.csv
└── index.qmdTo get from 01-cleaning.qmd we need to provide a relative path to where we are saving the file, including the folder name and the name of the file: "data-processed/01-hot100.rds".
We’ll need to do a similar thing when import data in the next notebook.
3.10 Outline and bookmarks
I didn’t want to break our flow of work to explain this earlier, but I want you show you some nice features in RStudio to jump up and down your notebook.
- At the top-right of your document window is a button called Outline. This recognizes your Markdown headlines and allows you to jump up and down your document by clicking on them.
- At the bottom of your window above the Console you’ll see a dropdown box that shows the main headlines and code chunks in your notebook. You can also use those to move up and down your notebook.
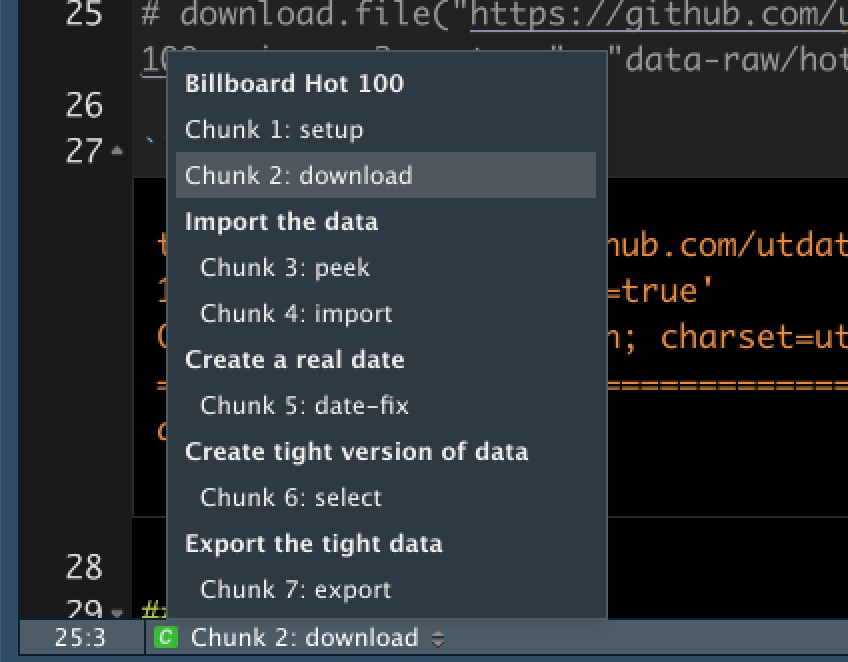
You’ll notice that some of my code chunks also have names, but yours probably don’t. It’s optional to name chunks with a label but it helps you find them through the bookmarks. In addition, plots produced by the chunks will have useful names that make them easier to reference elsewhere, but that’s a lesson for later in the semester.
Here is how you can name a chunk by using “execution options”, which can control your code output. (This example shows all of the R code chunk.)
```{r}
#| label: select-rows
hot100_clean |> select(title, performer) |> head()
```Your chunk labels should be short but evocative and should not contain spaces. Hadley Wickham recommends using dashes - to separate words (instead of underscores _) and avoiding other special characters in chunk labels.
You can read more execution options and chunks in R for Data Science.
3.11 Render and publish
Lastly, I want you to render your notebook so you can see the pretty HTML version.
- Click the Render button in the toolbar (or use Cmd-Shift-k).
This should open the HTML version of your page in the Viewer pane. Note there is also a button in that View toolbar that will instead open your site in your web browser.
But note this version is only available on your computer. In a minute we’ll publish this to Quarto Pub so you have an online version that we’ll continue to update.
3.11.1 Publish to Quarto pub
Now the we have that in order, let’s publish this to the internet. If you published your last project as instructed, it should remember your credentials.
- In the bottom-left pane of RStudio, click on the pane Terminal.
- Type in
quarto publishand hit Return. - The first option available (and perhaps the only) should be Quarto Pub. Hit Return to choose that.
- It might know who you are already, but continue through the prompts.
You should end up with your browser open to your new Quarto Pub website.
Well done.
3.12 Review of what we’ve learned so far
Most of this lesson has been about importing and cleaning data, which can sometimes be the most challenging part of a project. Here we were working with well-formed data, but we still used quite a few tools from the tidyverse like readr (read_csv, write_rds) and dplyr (select, mutate).
Here are the functions we used and what they do. Most are linked to documentation sites:
library()loads a package so we can use functions within it. We will use at leastlibrary(tidyverse)in every project.read_csv()imports a csv file. You want that function with the underscore, notread.csv().clean_names()is a function in the janitor package that standardizes column names.head()prints the first 6 rows of your data unless you specify differently within the function.glimpse()is a view of your data where you can see all of the column names, their data type and a few examples of the data.summary()gives you some quick summary statistics about your data like min, max, mean, median.mutate()changes data. You can create new columns or overwrite existing ones.mdy()is a lubridate function to convert text into a date. There are other functions depending on the way your text is ordered.arrange()sorts your data based on values within the variables you specify. Adddesc()to the variable if you want it in descending order.select()selects columns from your tibble. You can list all the columns to keep, or use!to remove columns. There are many variations.write_rds()writes data out to a file in a format that preserves data types.
You can find a list of all the functions used in this book, in the Functions chapter.
3.13 What’s next
This is part one of a two-chapter project. You might be asked to turn in your progress, or we might wait until both parts are done. Check your class assignments in Canvas.
Please reach out to me if you have questions on what you’ve done so far. These are important skills you’ll use on future projects.
3.6.3 Comment the download code
Now that we’ve downloaded the data to our computer, we don’t need to run this line of code again unless we want to update our data from the source. We can “comment” the code to preserve it but keep it from running again if we re-run our notebook (and we will).
download.file()function, then go under the Code menu to Comment/Uncomment Lines. (Note the keyboard command there Cmd-Shift-C, as it is another useful one!)Adding comments in programming is a common thing and every programming language has a way to do it. It is a way for the programmer to write notes to their future self, colleagues or — like in this case — comment out some code that you want to keep, but don’t want to execute when the program is run.
We write most of our “explaining” text outside of code chunks using a syntax called Markdown. Markdown is designed to be readable as written, but it is given pretty formatting when “printed” as a PDF or HTML file.
But, sometimes it makes more sense to explain something right where the code is being executed. If you are inside a code chunk, you start the comment with one or more hashes
#. Any text on that line that follows won’t be executed as code.Yes, it is confusing that in Markdown you use hashes
#to make headlines, but in code chunks you use#to make comments. We’re essentially writing in two languages in the same document.